¶ 1. Using Python with Conda
¶ 1.1 Command to using Conda
- Run command
source /data/programs/anaconda3/bin/activate - Run command
conda init --all - Logout and Login again
[peeranon@slurmmaster ~]$ source /data/programs/anaconda3/bin/activate
(base) [peeranon@slurmmaster ~]$ conda init
no change /data/programs/anaconda3/condabin/conda
no change /data/programs/anaconda3/bin/conda
no change /data/programs/anaconda3/bin/conda-env
no change /data/programs/anaconda3/bin/activate
no change /data/programs/anaconda3/bin/deactivate
no change /data/programs/anaconda3/etc/profile.d/conda.sh
no change /data/programs/anaconda3/etc/fish/conf.d/conda.fish
no change /data/programs/anaconda3/shell/condabin/Conda.psm1
no change /data/programs/anaconda3/shell/condabin/conda-hook.ps1
no change /data/programs/anaconda3/lib/python3.11/site-packages/xontrib/conda.xsh
no change /data/programs/anaconda3/etc/profile.d/conda.csh
modified /data/users/peeranon/.bashrc
==> For changes to take effect, close and re-open your current shell. <==
(base) [peeranon@slurmmaster ~]$ logout
[root@slurmmaster anaconda]# su - peeranon
Last login: Tue May 7 20:29:31 +07 2024 on pts/0
(base) [peeranon@slurmmaster ~]$ conda env list
# conda environments:
#
base * /data/programs/anaconda3
(base) [peeranon@slurmmaster ~]$
¶ 1.2 Creating Virtual Environment on Conda
Create Virtual Environment on Conda ด้วสั่ง conda create -n <name>
(base) [peeranon@slurmmaster ~]$ conda create -n python3.6 python=3.6
Channels:
- defaults
Platform: linux-64
Collecting package metadata (repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /data/users/peeranon/.conda/envs/python3.6
added / updated specs:
- python=3.6
The following packages will be downloaded:
package | build
---------------------------|-----------------
ca-certificates-2024.3.11 | h06a4308_0 127 KB
certifi-2021.5.30 | py36h06a4308_0 139 KB
libffi-3.3 | he6710b0_2 50 KB
openssl-1.1.1w | h7f8727e_0 3.7 MB
pip-21.2.2 | py36h06a4308_0 1.8 MB
python-3.6.13 | h12debd9_1 32.5 MB
setuptools-58.0.4 | py36h06a4308_0 788 KB
sqlite-3.45.3 | h5eee18b_0 1.2 MB
tk-8.6.14 | h39e8969_0 3.4 MB
wheel-0.37.1 | pyhd3eb1b0_0 33 KB
xz-5.4.6 | h5eee18b_1 643 KB
zlib-1.2.13 | h5eee18b_1 111 KB
------------------------------------------------------------
Total: 44.5 MB
The following NEW packages will be INSTALLED:
_libgcc_mutex pkgs/main/linux-64::_libgcc_mutex-0.1-main
_openmp_mutex pkgs/main/linux-64::_openmp_mutex-5.1-1_gnu
ca-certificates pkgs/main/linux-64::ca-certificates-2024.3.11-h06a4308_0
certifi pkgs/main/linux-64::certifi-2021.5.30-py36h06a4308_0
ld_impl_linux-64 pkgs/main/linux-64::ld_impl_linux-64-2.38-h1181459_1
libffi pkgs/main/linux-64::libffi-3.3-he6710b0_2
libgcc-ng pkgs/main/linux-64::libgcc-ng-11.2.0-h1234567_1
libgomp pkgs/main/linux-64::libgomp-11.2.0-h1234567_1
libstdcxx-ng pkgs/main/linux-64::libstdcxx-ng-11.2.0-h1234567_1
ncurses pkgs/main/linux-64::ncurses-6.4-h6a678d5_0
openssl pkgs/main/linux-64::openssl-1.1.1w-h7f8727e_0
pip pkgs/main/linux-64::pip-21.2.2-py36h06a4308_0
python pkgs/main/linux-64::python-3.6.13-h12debd9_1
readline pkgs/main/linux-64::readline-8.2-h5eee18b_0
setuptools pkgs/main/linux-64::setuptools-58.0.4-py36h06a4308_0
sqlite pkgs/main/linux-64::sqlite-3.45.3-h5eee18b_0
tk pkgs/main/linux-64::tk-8.6.14-h39e8969_0
wheel pkgs/main/noarch::wheel-0.37.1-pyhd3eb1b0_0
xz pkgs/main/linux-64::xz-5.4.6-h5eee18b_1
zlib pkgs/main/linux-64::zlib-1.2.13-h5eee18b_1
Proceed ([y]/n)? y
¶ 1.3 UsingConda Virtual Environment
Run command conda activate <package_name> to show Environment in the left hand of the line such as (base) or (python3.6)
(base) [peeranon@slurmmaster ~]$ python -V
# conda environments:
#base) [peeranon@slurmmaster ~]$ conda env list
base * /data/programs/anaconda3
python3.6 /data/users/peeranon/.conda/envs/python3.6
(base) [peeranon@slurmmaster ~]$ conda activate python3.6
(python3.6) [peeranon@slurmmaster ~]$ python -V
Python 3.6.13 :: Anaconda, Inc.
(python3.6) [peeranon@slurmmaster ~]$
¶ 1.4 Installing Library in Conda Virtual Environment
Run command conda install <package_name>
(python3.6) [peeranon@slurmmaster ~]$ conda install pandas
¶ 1.5 Creating Conda Environment to use on Jupyter Notebook
- Activate your environment
conda activate python3.6 - Install library Jupyter Notebook package
conda install notebook - Install library ipykernel package
conda install ipykernel - Run command
python -m ipykernel install --user --name=<any_name_for_kernel> - Open Jupyter Notebook and follow the SSH tunnel in next section
- Verify Kernel on Jupyter Notebook
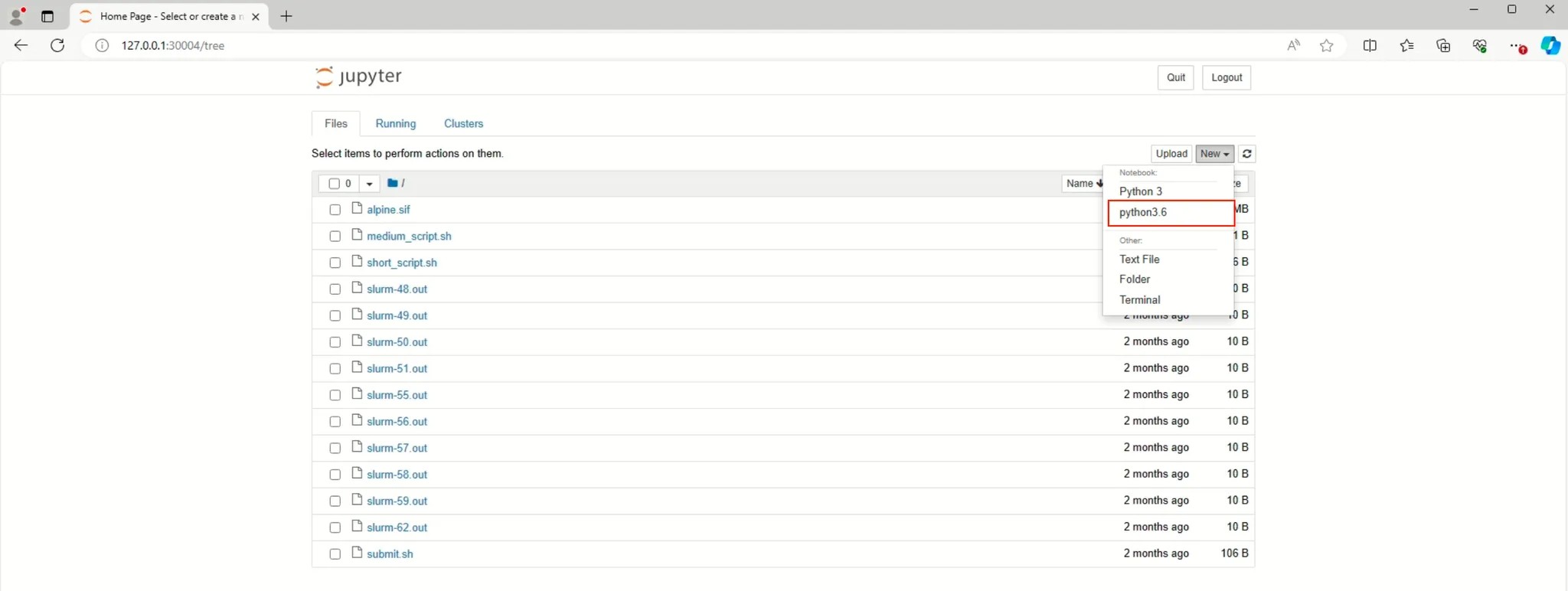
¶ 1.6 Delete Virtual Environment on Conda
Run command conda env remove --name <environment_name>
¶ 1.7 Deactivate Conda Virtual Environment
Run command conda deactivate
¶ 2. Running Jupyter Notebook job on SLURM Workload Manager and using SSH Tunnel
Users can run Jupyter Notebook jobs on compute nodes and access them from their own machines or laptop, with Slurm managing resource allocation for these jobs.
Initially, Jupyter Notebook has already been installed within a Conda environment.
¶ 2.1 Running Jupyter Notebook job on SLURM Workload Manager
Consists of 3 steps:
2.1.1) Start Jupyter Notebook Batch Job on Slurm
2.1.2) Start SSH Tunnel
2.1.3) Access the notebook through a browser on the user's machine or laptop
¶ 2.1.1) Start Jupyter Notebook Batch Job ผ่าน Slurm
Below is an example script for submitting a Jupyter Notebook job as a batch job (the sample file is located at /data/examples/notebook.sh). Users should copy it to their home and adjust the Slurm settings (#SBATCH) as appropriate for their use case.
Once the script has been modified, submit the job using the following command:sbatch /data/examples/notebook.sh
#!/bin/bash
#SBATCH --partition gpu
#SBATCH --mem-per-cpu 8G
#SBATCH --time 1-0:00:00
#SBATCH --job-name jupyter-notebook
#SBATCH --output jupyter-notebook-%J.log
# get tunneling info
XDG_RUNTIME_DIR=""
port=$(shuf -i8000-9999 -n1)
node=$(hostname -s)
user=$(whoami)
cluster=$(hostname -f | awk -F"." '{print $2}')
# print tunneling instructions jupyter-log
echo -e "
MacOS or linux terminal command to create your ssh tunnel
ssh -N -L ${port}:${node}:${port} ${user}@${cluster}
Windows MobaXterm info
Forwarded port:same as remote port
Remote server: ${node}
Remote port: ${port}
SSH server: ${cluster}
SSH login: $user
SSH port: 22
Use a Browser on your local machine to go to:
localhost:${port} (prefix w/ https:// if using password)
"
# load modules or conda environments here
# uncomment the following two lines to use your conda environment called notebook_env
# module load miniconda
# source activate notebook_env
# DON'T USE ADDRESS BELOW.
# DO USE TOKEN BELOW
jupyter-notebook --no-browser --port=${port} --ip=${node}
(base) [peeranon@slurmmaster ~]$ sbatch /data/examples/notebook.sh
Submitted batch job 169
¶ 2.1.2) Start SSH Tunnel
Once the job has been submitted, the Jupyter Notebook server will start and be ready for connection. Users can check their running jobs by executing the command:squeue -u $USER
A status of R in the ST column indicates that the job is currently running. If the status is PD, please wait a moment until the job starts before attempting to connect.
The log file containing the connection details will be named:jupyter-notebook-[jobid].log where jobid refers to the job ID assigned by Slurm
(base) [peeranon@slurmmaster ~]$ squeue -u peeranon
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
169 gpu jupyter- peeranon R 7:50 1 ai3
Below is example of log file that describe how to use Jupyter Notebook
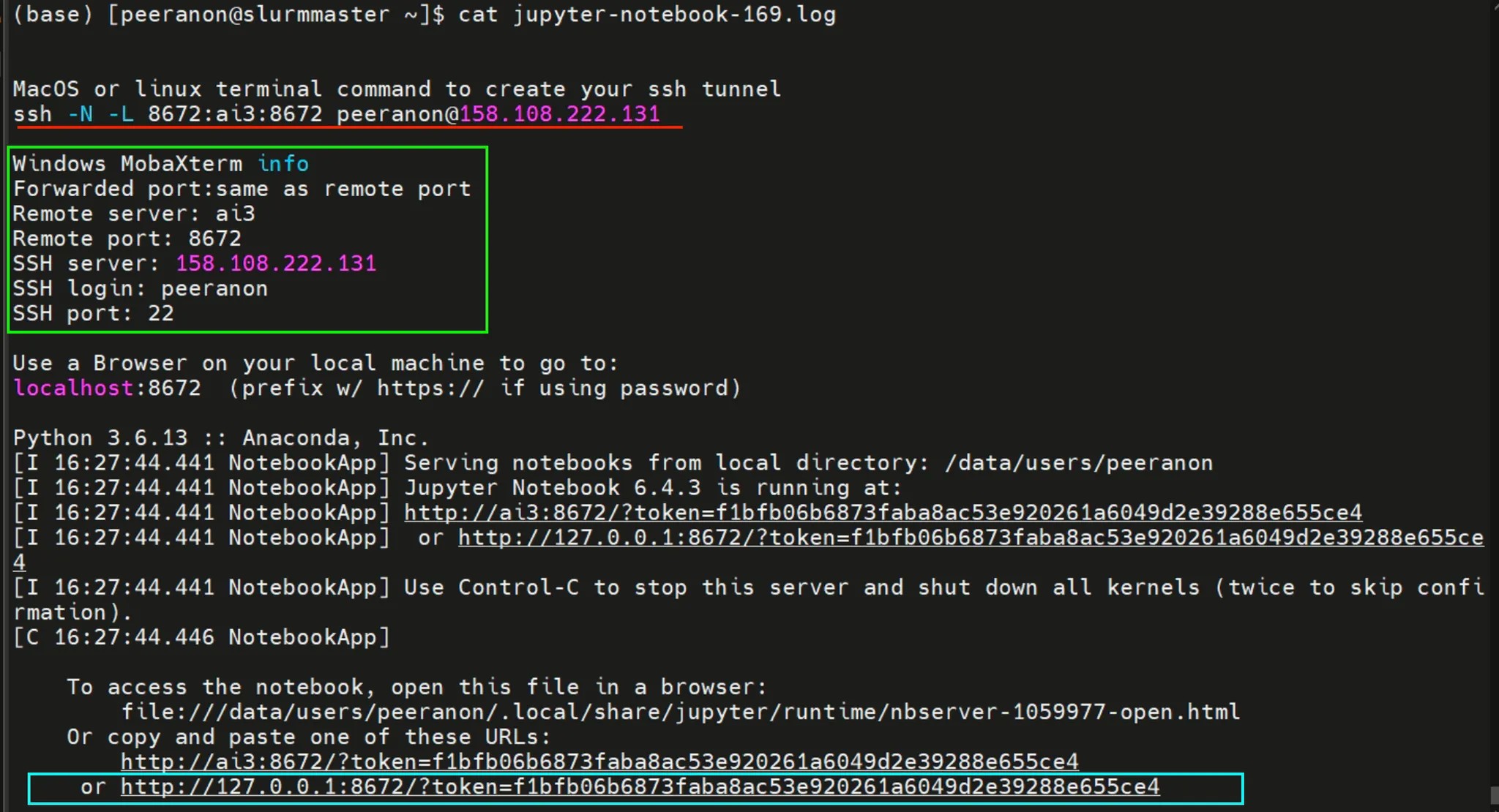
¶ SSH Tunnel on MacOC and Linux
For Mac OS and Linux users, open the Terminal and run the command shown in the below image. After running the command, you will be prompted to enter your password. Make sure to enter it correctly. If the password is entered successfully, you should see an output similar to the image below.

¶ SSH Tunnel on Windows
For Windows users, we recommend using MobaXterm. You can set up an SSH tunnel by following these steps:
- Click Start local Terminal
- Run command :
ssh -N -L 8672:ai3:8672peeranon@158.108.222.131 - Enter your password. If the password is entered successfully, you should see an output similar to the image below.

¶ 2.1.3) Access the Jupyter Notebook on user's machine or laptop
After successfully setting up the SSH tunnel, scroll to the end of the log file. You will find a URL that includes a token—this is your access link. Copy the entire URL (as shown in the blue box) and paste it into your browser to access the Jupyter Notebook.
<http://127.0.0.1:8672/?token=f1bfb06b6873faba8ac53e920261a6049d2e39288e655ce4>

Below is successful access Jupyter Notebook.
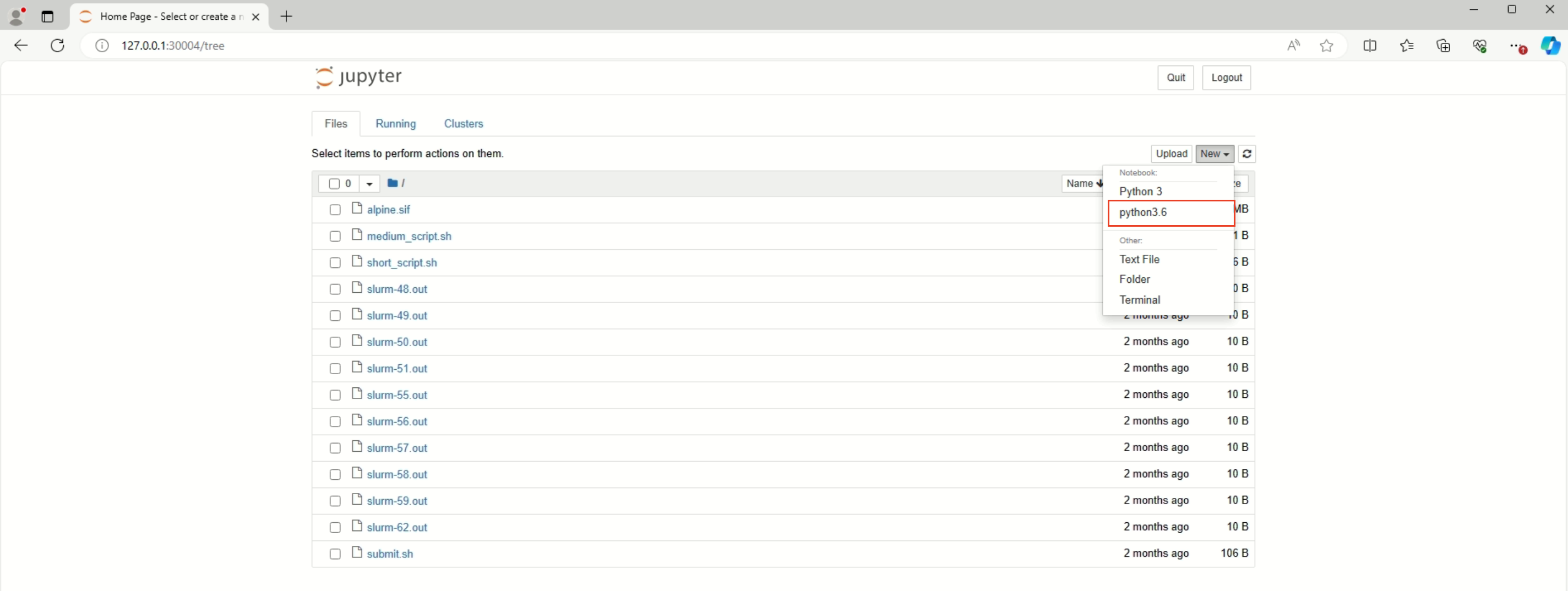
¶ 2.2 Stop Jupyter Notebook
To stop using the Jupyter Notebook, follow these steps:
- Close the web browser.
- Terminate the SSH tunnel.
- Cancel the running job on Slurm using the following command:
scancel -f <jobid>Replace<jobid>with your actual Slurm job ID.
(base) [peeranon@slurmmaster ~]$ squeue -u peeranon
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)
169 gpu jupyter- peeranon R 19:54 1 ai3
(base) [peeranon@slurmmaster ~]$ scancel -f 169
(base) [peeranon@slurmmaster ~]$ squeue -u peeranon
JOBID PARTITION NAME USER ST TIME NODES NODELIST(REASON)